The Role of Stochasticity in the Evolution of Animal Pattern and Morphology
Biophysics Internship - 2022
Cover image credit from Figure 1.16 of (Begon & Townsend, 2020).
Gene Regulatory Networks (GRNs) are systems of genes that interact with one another through inhibitions and activations to control gene expression. They play a central role in determining an organism’s morphology. As a result, variation of these networks lead to evolutionary changes in morphology and so are of great use in simulating evolutionary processes.
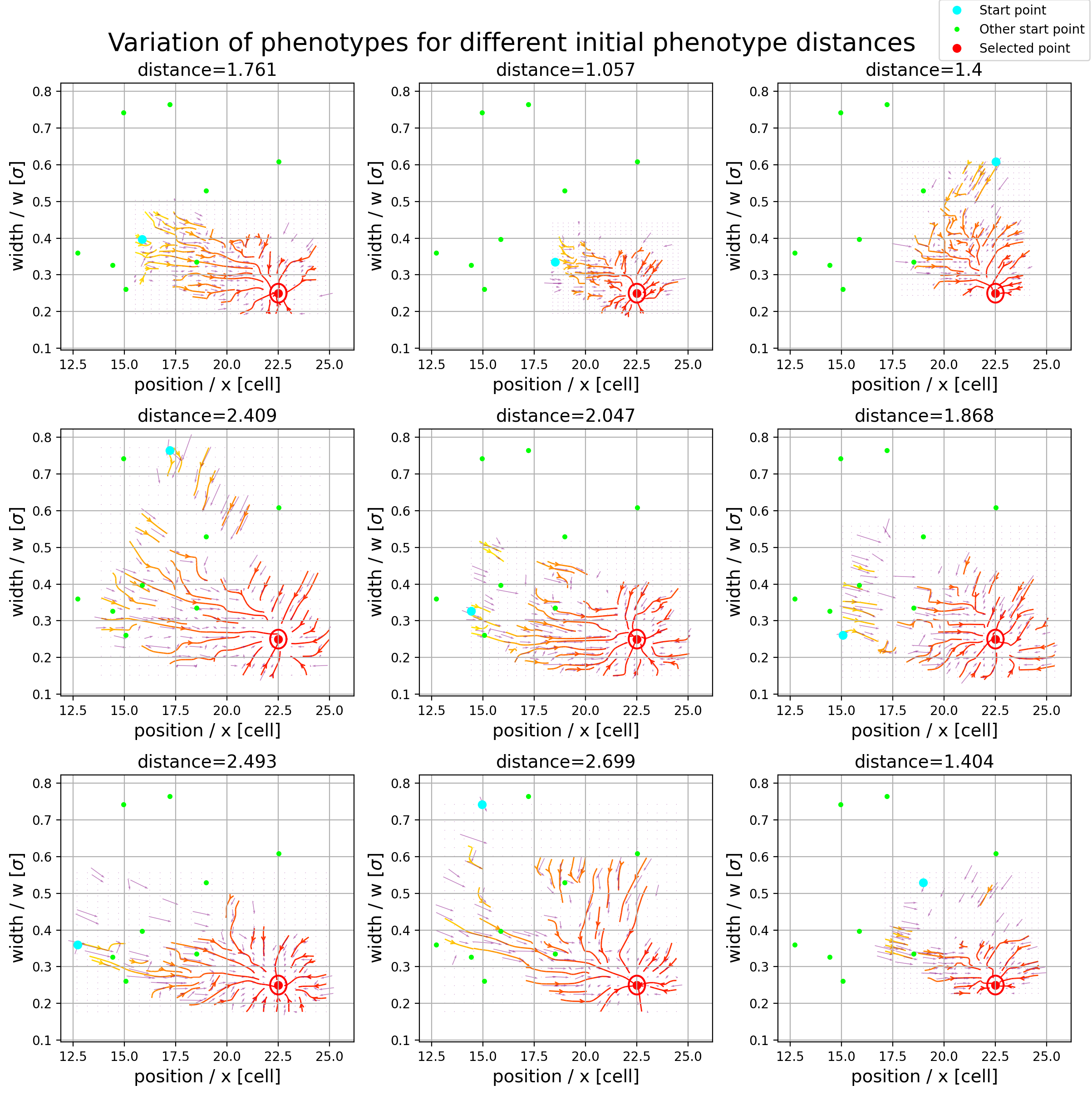
We used GRNs, specifically a three gene topology, to investigate convergence in evolution. To do this we solved the GRN for the steady state numerically using the coupled, non-linear, differential equations \begin{equation} \frac{dg_{ij}}{dt} = \phi\bigg(\chi\Big(\sum\limits_{l=1}^3 W^{il}g_{lj} + M\Big)\bigg) - \lambda g_{ij} \label{ODE} \end{equation} where $\chi$ is a heaviside function and $\phi$ is a Michaelis-Menten function. This equation encompasses the interaction between the genes in the summation over a column of weights, $W$, the morphogen input in $M$ and a degradation rate in $\lambda$. By rerunning the evolutionary process while selecting for the same phenotype we explored how the final genotypes varied and found correlations between the weights of the interactions between the genes. We found clear correlations between some of the interactions and also explored the ``flow’’ of the organism through the phenotype space in order to locate common routes evolution takes as can be seen in Figure 1. Further details can be found here.